Universal - Flex (2x) qPCR-Master for probes and novel ROX-alike reference dye
The Uni-qPCR Master mix offers quantitative PCR for probes with a new ROX-alike reference dye for using with all common PCR-Platforms.
The Universal-Flex qPCR Mastermix for probes is compatible across a variety of instrument platforms including those that require a high, low or no ROX reference signal
ready to use 2x PCR Master Mix for probes with Blue dye. 2 x 1 ml, two fold concentrated
Product Description
The Bio-Star 2X Universal-flex probe qPCR Master Mix is a reaction mix optimized for real-time qPCR detection and quantitation of target DNA sequences using hydrolysis probes. It contains Hot Start Taq DNA polymerase, dNTPs, MgCL2, reaction buffer additives and stabilizers, optimized for Probe qPCR to ensures:
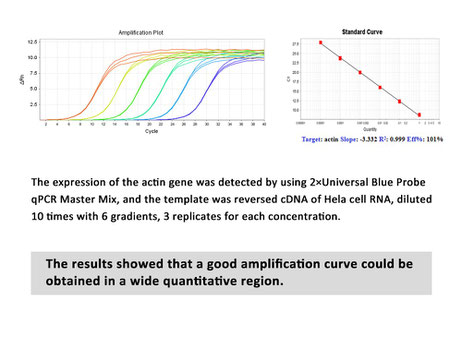
- a perfect amplification curve
- accurate quantification of target genes
- increased PCR specificity and sensitivity
- good repeatability and high reliability
- over a broad dynamic range.
Only templates, primers, probes and Nuclease-free Water need to be added for use. It also features a unique passive reference dye that is compatible across a variety of instrument platforms and a non-fluorescent, visible dye to monitor reaction setup. This dye does not spectrally overlap fluorophores commonly used for qPCR and will not interfere with real-time detection.
Taqs: Luna, NEB, Universal, quantitation of target DNA and cDNA sequences.
Final price excl. shipping costs3
- verfügbar / avaílable
- 1 - 3 days for delivery / 1 - 3 Tage Lieferzeit1
Storage and
transportation: at -20 °С;. Shipping
with blue ice or at room temperature
Storage
terms: up to 18 months
The Universal-Flex qPCR Mastermix for probes is compatible across a variety of instrument platforms including those that require a high, low or no ROX reference signal
Universal qPCR Mastermix Amplification protocol
1. Defrost the reaction mixture and stir thoroughly.
2. Add the following components into the thin-wall PCR tubes considering the final volume of a reaction mixture equal to 50 μl:
| Component | Volume | Final concentration |
| 2x Mastermix | 25 µl | 1x |
| Forward Primer *1 | variable | 0,2 µM |
| Reverse Primer *1 | variable | 0,2 µM |
| DNA Template *2 | variable | 10 pg - 1 µg |
| Sterile Water | up to 50 µl |
3. Gently vortex and remove droplets by centrifugation.
4. Perform PCR
*1: a: Usually, a good amplification effect can be obtained with the final concentration of 0.2 μM. When the reaction performance is poor, the primer concentration can be adjusted in the range of 0.2-1.0 μM.
*2: b: The amount of template added varies depending on the number of copies of the target gene, and the appropriate amount of template addition is studied by gradient dilution. The best addition amount of template DNA in the 20 µl reaction system was less than 100 ng.
Universal qPCR Mastermix Cycler program
| Step |
Temp. °C |
Incubation time
|
Number of Cycles |
|
Preliminary denaturation |
95 | 0,5-2 min | 1 |
|
Denaturation |
95 | 15 sec | 30-40 |
|
Annealing |
55-65 |
10 sec |
30-40 |
| Elongation | 72 | 30 sec | 30-40 *1 |
|
Melting curve (recommended) |
1 | 1 |
as an alternative:
| Step |
Temp. °C |
Incubation time
|
Number of Cycles |
|
Preliminary denaturation |
95 | 0,5-2 min | 1 |
|
Denaturation |
95 | 15 sec | 30-40 |
|
Annealing / Extension |
60 |
30 sec |
30-40 *1 |
| Elongation | 72 | 30 sec | 30-40 |
|
Melting curve (recommended) |
1 | 1 |
*1: If amplification specificity needs to be improved, two-step procedure or annealing temperature can be used; To improve the amplification efficiency, a three-step procedure or extension time can be used.
Compatible instruments / Cycler List
ABI: 5700, 7000, 7300, 7700, 7900, 7900HT, 7900 HT Fast, StepOne™, StepOne Plus™, 7500/7500 Fast, ViiA 7™,;
Analytik Jena: qTOWER series;
qTOWER: LineGene series
Stratagene: Mx3000P®, 3005P™, 4000™;
Bio-Rad: CFX96™, CFX384™, iCycler iQ™, iQ5™, MyiQ™, MiniOpticon™, Opticon®, Opticon 2, Chromo4™;
Eppendorf: Realplex 2s, Mastercycler® ep, Realplex;
IIIumina: Eco QPCR;
Cepheid: SmartCycler®; QuantStudio™ series, PikoRealTM Cycler
Qiagen Corbett: Rotor-Gene® series;
Roche: LightCycler™ series;
Takara: Thermal Cycler Dice series;
Related products
PCR MASTERMIX SELECTION TABLE
PCR Mastermixes with dTTP and SybrGreen
BioStar 2 PCR mastermix with SYBRGreen, blue dye, no
rox
BioStar 3 PCR mastermix with SYBRGreen, 60 nm LOW-ROX
BioStar 4 PCR mastermix with SYBRGreen, 900 nm HIGH-ROX
PCR Mastermixes with dTTP for probes
BioStar 5 PCR mastermix for probes
BioStar 6 PCR mastermix for probes 60 mM LOW-ROX
BioStar 7 PCR mastermix for probes 900 mM HIGH-ROX
PCR Master Mixes with Evagreen and dUTP
PCR Master Mix for qPCR EVA 1 dUTP+ EvaGreen
PCR Master Mix for qPCR EVA 2 +UNG
PCR Master Mix for qPCR EVA3 +ROX
PCR Master Mix for qPCR EVA4 +ROX+UNG
PCR Master Mix for qPCR EVA5 +low ROX
PCR Master Mix for qPCR E6 +low ROX+UNG
PCR Master Mixes for probes and dUTP
qPCR Master Mix DLP1 Exclusiv +dUTP
qPCR Master Mix DLP2 Exclusiv +dUTP+UDG
qPCR Master Mix DLP3/DLP5 Exclusiv ROX (500 nM, 100nM)
qPCR Master Mix DLP4/DLP6 Exclusiv +ROX 500nM or 100 nM+UDG
Deutsche Beschreibung

Bio-Star Universal-Flex qPCR-Master Mix for probes, blue dye and a novel ROX-likely reference dye
Bio-Star Universal-Flex qPCR-Mastermix for probe; Blue dye and a novel ROX-likely reference dye
- accurate quantification of target genes
- increased PCR specificity and sensitivity
- good repeatability and high reliability
- over a broad dynamic range.
Material Safety Datasheet

References / Protocols / Notes / Recomendations / Tests
Taqman primer design principle
1. Determine the probe before designing primers.
2. When designing primers, get as close to the probe as possible without overlapping the probe.
3. Avoid using 4 or more consecutive G.
4. The Tm value of each primer should be 58-60°C.
5. The last 5 nucleotides at the end of the primer cannot have more than 2 G and C.
6. Primers had better not contain self-complementary sequences, otherwise they will form the hairpins.
7. In order to avoid the amplification of the genome, it is best to design primers across exons.
8. The length of amplification product should be 50-150 bp in order to obtain the best PCR efficiency.
9. No other non-specific products were found in the comparison results on NCBI.
Taqman probe design principle
1. Probe length should be 13-25 bp (13-30 bp if conventional TaqMan probe is used).
2. The Tm value should be 65°C~70°C, which is usually 5°C~10°C higher than the TM value of the primer to ensure that the probe preferentially binds to the target gene during annealing.
3. For a primer, the content of guanine-cytosine (G+C) should be between 40% and 70%.
4. The 5 'end of the probe should avoid using G, because the 5' end G will have quenching effect, even if it is cut off.
5. In the whole probe, the content of C is obviously higher than that of G, and the high content of G will have quenching effect, so we can choose another paired chain as the probe.
Taqman MGB probe design principle
1. A report dye (for example, FAMTM) is attached to the 5 'end of the probe.
2. There is a non-fluorescence quenching group (NFQ) at the 3 'end of the probe.
3. The part of MGB is attached to NFQ, and MGBs increases the annealing temperature (Tm) without increasing the length of the probe, so a shorter probe can be designed, but not less than 13 bp.
4. In principle, as long as there is a base mutation in the MGB probe, MGB can detect it (the MGB probe will not bind to the target gene and will not produce a fluorescent signal).